The Study of Cryptic Diversity: From Field to Lab
Posted in Science on May 24 2011, by Vinson Doyle
| Vinson Doyle, Graduate Student in Plant Genomics |

Plants and fungi have an intimate relationship. Some fungi, like mycorrhizal fungi which help a plant’s roots use soil resources more efficiently, are beneficial to plants, while others, like powdery mildew on squash leaves, are obviously harmful. However there are many fungi that exist somewhere on the continuum between friend and foe. To further complicate the matter, whether an individual fungus is beneficial, neutral, or antagonistic may change over time such that apparently harmless fungi can become pathogenic.

Studying these fungi can be difficult; they do not always make their presence obvious. Imagine tracking an elephant through the forests of East Africa to understand what it eats, where it travels, and how it selects a mate. Now, imagine tracking an individual you can’t see in the wild, or if you can see it, you can’t tell it apart from its siblings. Tracking these wild fungi would have been impossible a decade ago, but with the advances being made in modern genetic methods both here at The New York Botanical Garden and around the world, we are finally able to address the big questions surrounding these tiny organisms.
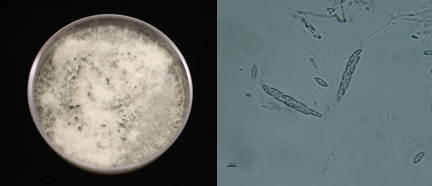
My work focuses on understanding the genetic diversity of a single fungal species, Colletotrichum gloeosporioides. C. gloeosporioides is a sneaky fungus that is capable of shifting its place on the continuum between harmless and pathogenic to cranberry. The main focus of my research involves understanding how the spores of C. gloeosporioides are dispersed, what other plants it is associated with, and how it reproduces. I hope that a fuller understanding of this fungus will help cranberry growers formulate effective management plans.
But before we can find answers we have to find the fungus.

My field work begins in the cultivated and wild cranberry bogs of North America in the hope that by finding the plant, we will find the fungus. In some cases, it is readily obvious that the plants have been infected by a pathogenic fungus. However, we frequently can’t determine which fungus has infected them, so we collect fruits, stems, and leaves and bring them back to the lab for further study. In other cases, we find plants that look perfectly healthy, but we suspect there are fungi lurking beneath the surface. So we take the material back to the Pfizer Plant Research Laboratory for more research. The hunt continues!
In order to study plant-associated fungi, that is, fungi that live within a plant such as C. gloeosporioides does, we must first coax the fungi out of the plant. To do this we place the plant pieces on a suitable growth medium that encourages the fungus to emerge from the plant and to populate the medium; in this way we are able to isolate the fungus and study it more carefully. Inevitably, many different species of fungi emerge onto the growth medium, which means we must isolate and identify each one.

Once we have isolated all the individual fungal strains, we use methods similar to those used in forensics to identify each one. In the Cullman Lab, we use DNA markers that allow us to identify each individual fungal strain within a single species in the same way that forensics specialists use DNA markers to establish whether an individual was at the scene of a crime.
Modern genetic methods have helped us determine that fungal strains are likely moving undetected (disguising themselves as harmless fungi) inside cranberry vines used to establish new farms in disparate regions before revealing themselves as harmful pathogens. This finding will hopefully allow researchers and farmers in the future to better understand the sources of disease epidemics. This knowledge should help farmers implement preventative measures and may lead to new methods for establishing cranberry bogs.
